paired end sequencing insert size
What is the optimal insert size. Mean 29622 STD2251 Read span.

What Are Paired End Reads The Sequencing Center
Here we modify the Illumina RNA ligation protocol to allow SS-PE sequencing by using a custom pre-adenylated 3 adaptor.

. Using the latest Illumina platform the MiSeq paired end reads of. Standard paired-end libraries 200500 bp can be used to detect large and small insertions deletions indels inversions and other rearrangements. The maximum distance x for a pair considered to be properly paired SAM flag 02 is calculated by solving equation Phix-musigmaxLp0 where mu is the mean sigma is the standard error of the insert size distribution L is the length of the genome p0 is prior of anomalous pair and Phi is the standard cumulative distribution function.
Here I wrote a simple script to search Cufflinks outputs. For Illumina systems DNA insert size range of 200800 bp. Java -jar picardjar CollectInsertSizeMetrics.
For the first test I took some sequence from the human genome hg19 and created two 100 bp reads from this region. If processing a small file set the minimum percentage option M to 05 otherwise an error may occur. These reads can then be sequenced using the same SP1-SP2 adapter protocols used in PE sequencing.
The reads were then mapped back to the reference using BWA aln and sampe. I am planning to perform RNA seq using a MiSeq Reagent Kit v3 600 cycle mean insert size of 600bp 2x 300bp reads paired-end. The fragment size which you need to select for during a gel purification for example would be the insert size length of both adapters around 120 bp extra for both Illumina.
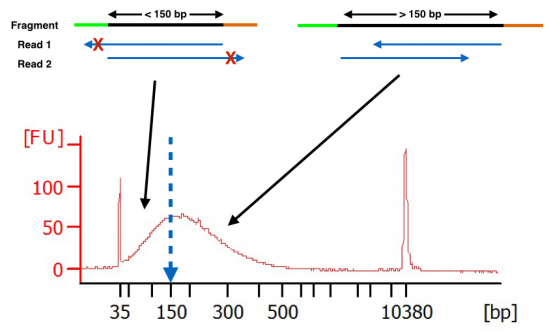
However some paired-end sequencing data show the presence of a subpopulation of reads where the second read R2 has lower average qualities. The reads have a length of typically 50 - 300 bp. The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle piece.
2Align paired-end RNA-Seq data to the reference transcripts using minimum insert-length as zero and maximum insert-length as 500 or more. The purpose of this protocol is to add adapter sequences onto the ends of DNA fragments to generate the following sequencing library format. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert size.
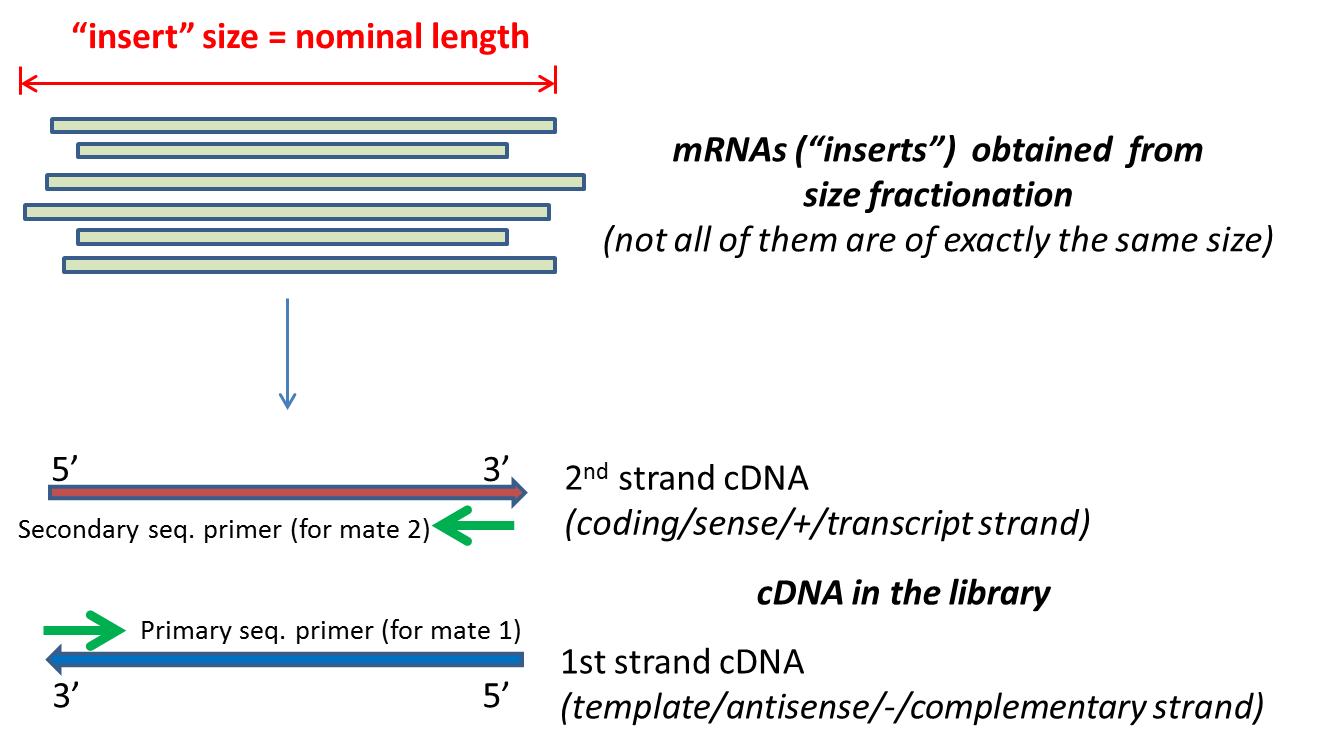
The new version of Cufflinks tries to estimate the mean and std value of paired-end read insert size automatically. Figure 1 Sequencing Library after PairedEnd Sample Preparation. The variation of insert sizes is often large and the average size difficult to control.
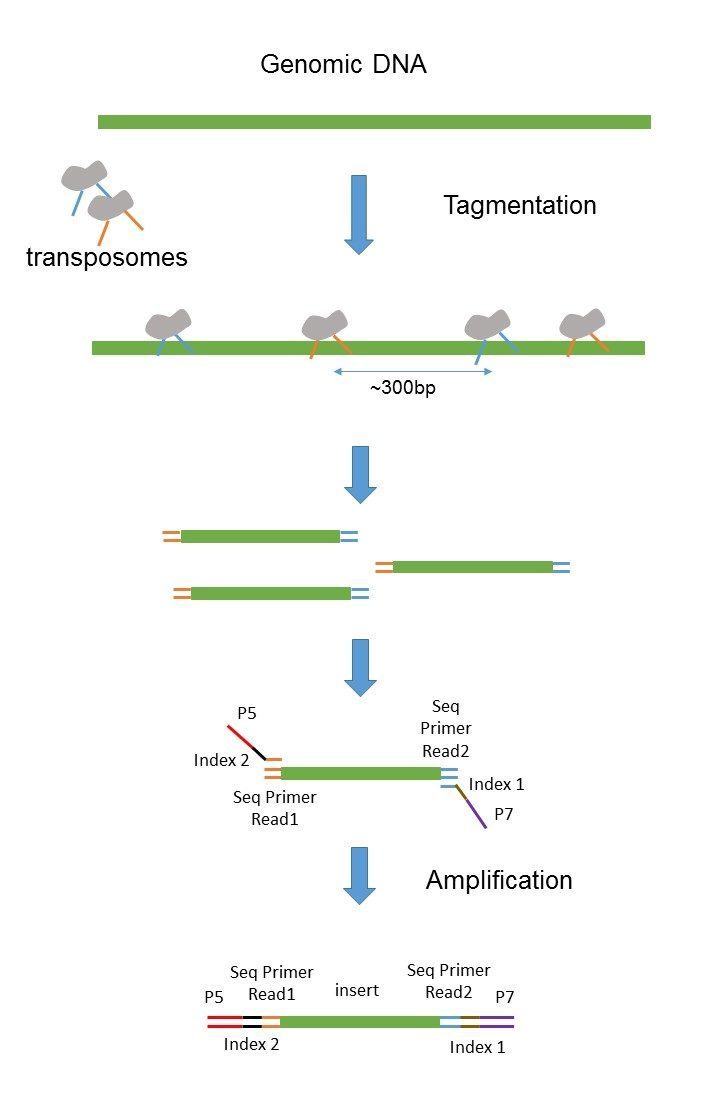
The implementation is easy. Please see InsertSizeMetrics for detailed explanations of each metric. The library preparation for paired-end sequencing according to the lllumina technology consists in fragmenting the genomic DNA mechanically Covaris Bioruptor or enzymatically tagmentase to sizes below 1 kb.
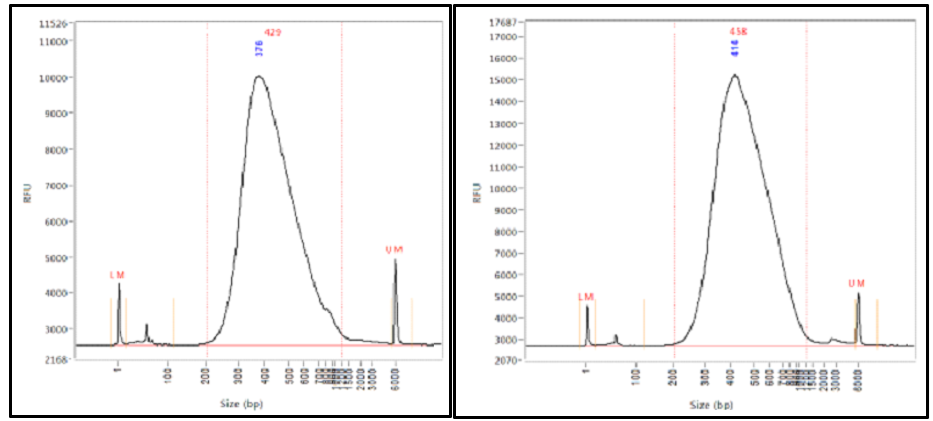
This can result in a proportion of fragments with an insert size of less than the length of a single read. Including flanking intronic regions of diagnostic significance eg due to splice variants of about 30bp at either side the average target of interest is about 220 bp long. Mean 48454 STD10836 Does it mean my insert size is 484-296184.
Sometimes we need to extract such information to check our sequencing data. Typically Illumina paired end reads have an insert longer than the combined length of both reads see Figure Figure1 1. To read more about the different parts of a prepared DNA fragment please take a look on the figure in the article about observed sequence lengths in Illumina.
Nine percent of read pairs were found to have an insert size of less than 250 bp and so likely to contain adapter after mapping to the reference genome of which 98 were also found to have an insert size of less than 250 bp by overlapping the reversed sequence. Sequencing on the Illumina sequencing platform. I got this output.
The inner mate distance between the two reads is 200 bp creating an insert size of 400 bp. Indexed and complementary adapters of the sequencing primers are then ligated at each end of the DNA fragments to ensure. Normally the insert size is longer than the sum of the two read lengths meaning there is an unsequenced inner part in the middle of the insert.
PE reads generally have a smaller insert size 1kp than MP 2-5 kb. However the average size of human coding exons is only 160bp. For instance for a PE150 sequencing run the insert size should be above 300bp.
Here we modify the Illumina RNA ligation protocol to allow SS-PE sequencing by using a custom pre-adenylated 3 adaptor. It depends on the used Illumina system the used reagents kits and the mode singlepaired end. Search for key words like.
Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert size. You can use sequencing reagents to generate single continuous reads or for paired-end sequencing in both directions. 1Create reference transcriptome index for BowTie or BWA.
Read length of paired-end reads. So why you get reads of the same length when you sequence. Now lets get started.
Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert size. The difference in insert size stems from the difference in protocols. For example a 300-cycle kit can be used for a 1 300 bp single-read run or a 2 150 bp paired-end run.
Insert size -- The insert size refers to the distance between the pairs. We show that the fragment length is a major driver of. The script is written in Python.
The sequencing starts at Read 1 Adapter mate 1 and ends with the sequencing from Read 2 Adapter mate 2. -I 0 -X 500 BWA. Paired-end sequencing also provides greater ability to overcome the obstacles of characterizing repetitive sequence ele-ments by filling in gaps of consensus sequence to achieve complete.
Paired End Sequencing France Genomique
How Short Inserts Affect Sequencing Performance
What Read Lengths Are Produced By Modern Illumina Sequencers

What Is Mate Pair Sequencing For

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram
What Happens When Hardware Advances Faster Than Molecular Protocols Cofactor Genomics

Pdf Assessment Of Insert Sizes And Adapter Content In Fastq Data From Nexteraxt Libraries
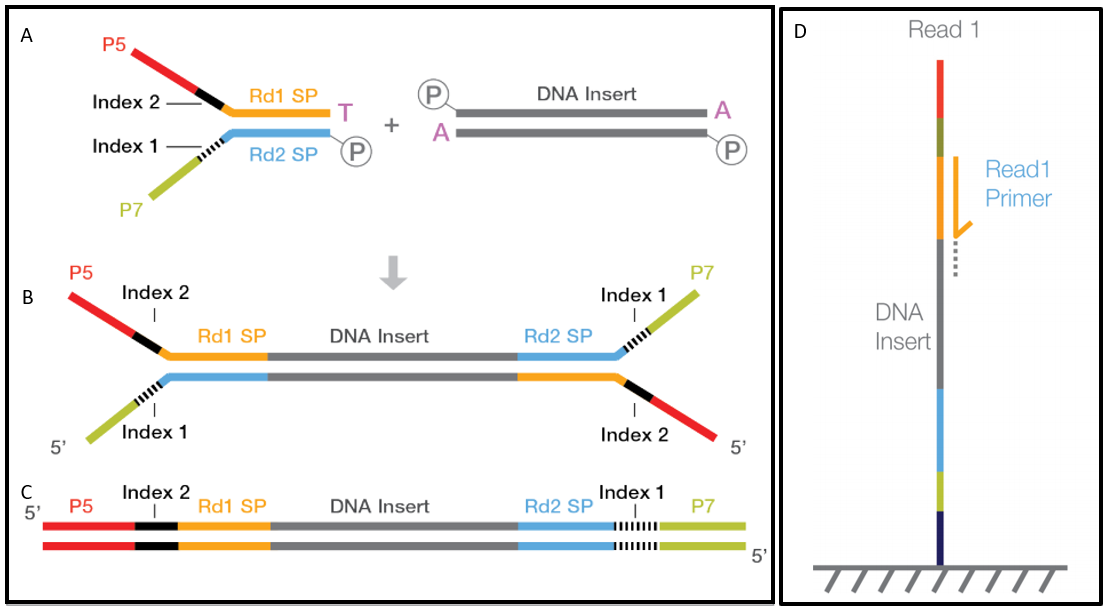
How Short Inserts Affect Sequencing Performance
Next Generation Sequencing Overview And Solutions To Common Problems

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram

What Are Paired End Reads The Sequencing Center

Interpreting Color By Insert Size Integrative Genomics Viewer
What Is Paired End 150 Omega Bioservices

Definition Of Essential Terms A Definition Of Fragment Read And Download Scientific Diagram
The Insert Size In Paired End Data Seqanswers

What Is Mate Pair Sequencing For

Interpreting Color By Insert Size Integrative Genomics Viewer